Principal Contact
Abstract:
Helicobacter pylori is a genetically diverse bacterial pathogen and its CagA gene is a major virulence factor that plays an important role in gastroduodenal pathologies. The biological function of cagA depends on tyrosine phosphorylation within the EPIYA (Glutamate-Proline-Isoleucine-Tyrosine-Alanine) motifs at the C-terminal region of the protein. This region may undergo polymorphism to give different types of EPIYA motifs. EPIYA motif diversity may provide a useful tool for prediction of H. pylori pathogenic activity and accurate determination of number and type of cagA EPIYA motifs could identify the virulent H. pylori. The aim of this study was to detect H. pylori cagA gene and its polymorphism in endoscopic gastroduodenal biopsy specimen from patients with gastroduodenal diseases in Bangladesh. This cross sectional study was carried out in the Department of Microbiology & Immunology, Bangabandhu Sheikh Mujib Medical University and Center for Advanced Research in Sciences, University of Dhaka during the period from March 2014 to February 2015. Gastric biopsies were collected from 78 patients with gastritis, duodenal ulcer, gastric ulcer and gastric carcinoma. H. pylori was identified by rapid urease test and ureC gene PCR. Presence of cagA gene and number and pattern of cagA EPIYA motif were determined by PCR. DNA sequencing was carried out to confirm the PCR detection method of cagA EPIYA motif and to analyse their peptide sequence. Among 31(39.7%) H. pylori positive cases, 19 (61.3%) were cagA gene positive in 11(55%) gastritis, 4(66.7%) duodenal ulcer, 2(66.7%) gastric ulcer and 2(100%) gastric carcinoma. A significant association was found between cagA gene and duodenal ulcer (p=˂0.05). EPIYA motif of all H. pylori cagA positive cases showed Western type cagA EPIYA ABC. No East Asian EPIYA ABD motif was found. Majority of gastroduodenal cases (57.9%) had 3 copies of EPIYA (ABC type), 26.3% had 4 copies (ABCC type) while remaining 10.5% had AC and 5.2% AB type EPIYA motif. EPIYA ABC was found in 75% of duodenal ulcer followed by 54.5% of gastritis and 50% of both gastric ulcer and gastric carcinoma patients. EPIYA ABCC motif was found in 50% of gastric ulcer and gastric carcinoma patients. Most of the EPIYA motif was EPIYA ABC and some were ABCC which has the risk of developing gastric carcinoma.
Key word: EPIYA (Glutamate-Proline- Isoleucine-Tyrosine- Alanine), polymorphism, PCR
Key word: EPIYA (Glutamate-Proline- Isoleucine-Tyrosine- Alanine), polymorphism, PCR
Introduction:
Helicobacter pylori is a spiral shaped microaerophilic bacterium that colonizes gastric mucosa of more than half of the world's population and is associated with development of complications such as peptic ulcer disease, gastric carcinoma and mucosa associated lymphoid tissue lymphoma.1 The association between H. pylori and gastric carcinoma led the World Health Organization to classify it as a class-I carcinogen in 1994. Bangladesh is a developing country and epidemiological studies shown 92% adult population were seropositive for H. pylori.2
Helicobacter pylori strains can be divided into two major types based on their ability to produce a 120–145kDa immune-dominant protein called cytotoxin associated gene A (CagA) antigen.3 H. pylori strains possessing the CagA gene were linked with an increased risk of developing gastric cancer and peptic ulcer. The risk of developing gastric cancer in H. pylori infected CagA-positive subjects is six fold higher than that in CagA-negative subjects.4 More than 90% of isolated strains from East Asia including Korea, Japan, and China are known to harbor cagA, while 50%-60% of isolated strains from Western countries are positive for it.5
The 3'-end region of cagA where the tyrosine phosphorylation sites are located are highly polymorphic.6 Phosphorylation occurs on specific tyrosine residues within repeating penta amino acid Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs, present at the C-terminus of the protein.7 Four different CagA EPIYA motifs, EPIYA-A, -B, -C, and -D have been defined based on the amino acid sequences surrounding the EPIYA residue.8 CagA proteins nearly always possess an EPIYA-A and an EPIYA-B, followed by various number of EPIYA-C repeats in Western-type or EPIYA-D motifs in East Asian type strains.6,9 It has been suggested that the considerable variation in number of repeating EPIYA-C or -D motifs determines the biological activity of cagA.7
CagA strains possessing multiple number EPIYA C segments predisposes to precancerous lesions and gastric cancer.7,10 Western type EPIYA ABC pattern present in 100% H. pylori strains isolated from patients with various gastroduodenal diseases in India.11 The phylogenetic analysis of the 5' end of the cagA gene indicates that Bangladeshi isolates are more closely related to H. pylori isolates from India and are different from isolates of East Asia.12
As large number of populations in Bangladesh are seropositive for H. pylori so, it is important to know the number and pattern of cagA EPIYA motifs for identifying the H. pylori infected patients who tend to develop severe gastroduodenal diseases. Therefore, EPIYA motif diversity may provide a useful tool for prediction of H. pylori pathogenic activity and accurate determination of number and type of cagA EPIYA motifs could identify the virulent H. pylori causing severe Gastroduodenal diseases in Bangladesh. So, this study was designed to detect H. pylori cagA gene polymorphism in gastric biopsy specimen from patients with gastroduodenal diseases.
Helicobacter pylori is a spiral shaped microaerophilic bacterium that colonizes gastric mucosa of more than half of the world's population and is associated with development of complications such as peptic ulcer disease, gastric carcinoma and mucosa associated lymphoid tissue lymphoma.1 The association between H. pylori and gastric carcinoma led the World Health Organization to classify it as a class-I carcinogen in 1994. Bangladesh is a developing country and epidemiological studies shown 92% adult population were seropositive for H. pylori.2
Helicobacter pylori strains can be divided into two major types based on their ability to produce a 120–145kDa immune-dominant protein called cytotoxin associated gene A (CagA) antigen.3 H. pylori strains possessing the CagA gene were linked with an increased risk of developing gastric cancer and peptic ulcer. The risk of developing gastric cancer in H. pylori infected CagA-positive subjects is six fold higher than that in CagA-negative subjects.4 More than 90% of isolated strains from East Asia including Korea, Japan, and China are known to harbor cagA, while 50%-60% of isolated strains from Western countries are positive for it.5
The 3'-end region of cagA where the tyrosine phosphorylation sites are located are highly polymorphic.6 Phosphorylation occurs on specific tyrosine residues within repeating penta amino acid Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs, present at the C-terminus of the protein.7 Four different CagA EPIYA motifs, EPIYA-A, -B, -C, and -D have been defined based on the amino acid sequences surrounding the EPIYA residue.8 CagA proteins nearly always possess an EPIYA-A and an EPIYA-B, followed by various number of EPIYA-C repeats in Western-type or EPIYA-D motifs in East Asian type strains.6,9 It has been suggested that the considerable variation in number of repeating EPIYA-C or -D motifs determines the biological activity of cagA.7
CagA strains possessing multiple number EPIYA C segments predisposes to precancerous lesions and gastric cancer.7,10 Western type EPIYA ABC pattern present in 100% H. pylori strains isolated from patients with various gastroduodenal diseases in India.11 The phylogenetic analysis of the 5' end of the cagA gene indicates that Bangladeshi isolates are more closely related to H. pylori isolates from India and are different from isolates of East Asia.12
As large number of populations in Bangladesh are seropositive for H. pylori so, it is important to know the number and pattern of cagA EPIYA motifs for identifying the H. pylori infected patients who tend to develop severe gastroduodenal diseases. Therefore, EPIYA motif diversity may provide a useful tool for prediction of H. pylori pathogenic activity and accurate determination of number and type of cagA EPIYA motifs could identify the virulent H. pylori causing severe Gastroduodenal diseases in Bangladesh. So, this study was designed to detect H. pylori cagA gene polymorphism in gastric biopsy specimen from patients with gastroduodenal diseases.
Materials and Methods:
Patients with dyspeptic symptoms who underwent endoscopic examination in the Department of Gastroenterology of Bangabandhu Sheikh Mujib Medical University (BSMMU) and Dhaka Medical College Hospital had endoscopic findings of any inflammation, ulcer or growth in gastroduodenal mucosa were enrolled in the study. Six biopsy specimens were collected from each patient: three from the gastric antrum and three from the gastric body.13 Each specimen from antrum and body was used for rapid urease test, PCR for ureC gene and histopathological examination. All procedures were performed in the Department of Microbiology and Immunology, BSMMU, Dhaka during the period of March, 2014 to February, 2015. Patients with history of previous gastric surgery, H. pylori eradication therapy or treatment with antibiotics, bismuth containing compounds, H2-receptor blockers or proton pump inhibitors within four weeks prior to enrolment were excluded from the study. Based on the endoscopic and histopatho-logical assessments, samples were divided into four groups of gastritis, duodenal ulcer, gastric ulcer and gastric carcinoma. Institutional Review Board, BSMMU and Ethical Review Committee of Dhaka Medical College approved the study protocol and informed consent was obtained from each patient prior to endoscopy and biopsy specimen collection. DNA extraction from gastric tissues: DNA from gastric tissues was extracted by using the QIAamp (QIAGEN) DNA Mini Kit according to the manufacturer's instruction. PCR assay: For confirming the presence of H. pylori DNA in gastric tissues, the ureC gene was identified by PCR using the primers14 (forward 5' AAGCTTTTAGGG GTGTTAGGGGTTT3' and reverse 5' AAGCTTACTTTCTAACACTAACG C 3' with a 294-bp size product. For detecting the presence of the cagA gene, PCR was performed by using primer pairs: forward (cagAF) 5'-GATAACAGGCAAGCTTTTGAGG-3' and reverse (cagAR) 5'-CTGCAAAAGATTGTTTG GCAGA -3' with a 349-bp size product.6 The forward primer cagA28F (5' TTCTCAAAGGAG CAATTGGC 3') and reverse primers cagAP1C (5'GTCCTGCTTTCTTTTTATTAACTTKAGC 3'), equimolar mixture of cagAP2CG (5'TTTAG CAACTTGAGCGTAAATGGG3') and cagAP2 TA(5'TTTAGCAACTTGAGTATAAATGGG3'), cagAP3E (5' ATCAATTGTAGCG TAAATG GG 3') and cagA-PD (5' TTGATTTGCCTCATC AA AATC 3') were used to amplify the EPIYA motif encoding the sequences A, B, C and D respectively.8 Amplification of the cagA3' variable region was performed using primers cag2(5' GGAACCCTAGTCGGTA ATG3') and cag4 (5'-ATCTTTGAGCTTG TCTATC G 3').8 All PCR in this study were performed in a volume of 25μl containing 10XPCR buffer, 500nM of each primer, 2mmol/L MgCl2; 200μM each deoxy nucleotide triphosphate (dNTPs), 1.5U Taq DNA polymerase and 200ng DNA sample. PCR was performed in a thermal cycler (Applied Biosystem 2720) under the following conditions: initial denaturation at 94°C for five minutes was followed by 35 cycles of 93°C for 1 minute, 55°C for 30 seconds, 72°C for 1 minute and final extension at 72°C for 10 minutes. The amplified products were analyzed in 2% agarose gel by electrophoresis. Following electro-phoresis, amplified product, the band of the sample was visualized under UV transilluminator. The bands were identified according to their molecular size by comparing with the molecular weight marker (100 bp DNA ladder) loaded in a separate lane. DNA sequencing: To confirm the type and number of EPIYA motif, direct sequencing of cagA 3´ variable region primer based amplified PCR product was performed. Sequencing was performed by Sanger dideoxy sequencing method at the Center for Advanced Research and Sciences, University of Dhaka. For automated DNA sequencing, PCR products were purified by using Thermo Scientific GeneJet PCR Purification Kit prior to cycle sequencing. The purified DNA was quantified and purities were evaluated by spectrophotometer (Nanodrop, ND 1000, Japan). After checking concentration of DNA, six samples were selected for sequencing. Purified products were sequenced using a BigDye Terminator Cycle Sequencing Kit in an ABI 3130 Genetic Analyzer (Applied Biosystems, USA). The sequences obtained were aligned using the CAP3 Sequence Assembly Program. After alignment, nucleotide sequences were transformed into amino acid sequences using the Blastx program and compared to previously published CagA gene sequence of strains H. pylori 26695 (AE000511) deposited into the GenBank.15 Statistical Analysis: After collection all data were checked, edited and analyzed by using computer based SPSS (Statistical package of social science) software. p value was calculated using Chi-square test to find the significant relationship. p value less than 0.05 was statistically significant.
Patients with dyspeptic symptoms who underwent endoscopic examination in the Department of Gastroenterology of Bangabandhu Sheikh Mujib Medical University (BSMMU) and Dhaka Medical College Hospital had endoscopic findings of any inflammation, ulcer or growth in gastroduodenal mucosa were enrolled in the study. Six biopsy specimens were collected from each patient: three from the gastric antrum and three from the gastric body.13 Each specimen from antrum and body was used for rapid urease test, PCR for ureC gene and histopathological examination. All procedures were performed in the Department of Microbiology and Immunology, BSMMU, Dhaka during the period of March, 2014 to February, 2015. Patients with history of previous gastric surgery, H. pylori eradication therapy or treatment with antibiotics, bismuth containing compounds, H2-receptor blockers or proton pump inhibitors within four weeks prior to enrolment were excluded from the study. Based on the endoscopic and histopatho-logical assessments, samples were divided into four groups of gastritis, duodenal ulcer, gastric ulcer and gastric carcinoma. Institutional Review Board, BSMMU and Ethical Review Committee of Dhaka Medical College approved the study protocol and informed consent was obtained from each patient prior to endoscopy and biopsy specimen collection. DNA extraction from gastric tissues: DNA from gastric tissues was extracted by using the QIAamp (QIAGEN) DNA Mini Kit according to the manufacturer's instruction. PCR assay: For confirming the presence of H. pylori DNA in gastric tissues, the ureC gene was identified by PCR using the primers14 (forward 5' AAGCTTTTAGGG GTGTTAGGGGTTT3' and reverse 5' AAGCTTACTTTCTAACACTAACG C 3' with a 294-bp size product. For detecting the presence of the cagA gene, PCR was performed by using primer pairs: forward (cagAF) 5'-GATAACAGGCAAGCTTTTGAGG-3' and reverse (cagAR) 5'-CTGCAAAAGATTGTTTG GCAGA -3' with a 349-bp size product.6 The forward primer cagA28F (5' TTCTCAAAGGAG CAATTGGC 3') and reverse primers cagAP1C (5'GTCCTGCTTTCTTTTTATTAACTTKAGC 3'), equimolar mixture of cagAP2CG (5'TTTAG CAACTTGAGCGTAAATGGG3') and cagAP2 TA(5'TTTAGCAACTTGAGTATAAATGGG3'), cagAP3E (5' ATCAATTGTAGCG TAAATG GG 3') and cagA-PD (5' TTGATTTGCCTCATC AA AATC 3') were used to amplify the EPIYA motif encoding the sequences A, B, C and D respectively.8 Amplification of the cagA3' variable region was performed using primers cag2(5' GGAACCCTAGTCGGTA ATG3') and cag4 (5'-ATCTTTGAGCTTG TCTATC G 3').8 All PCR in this study were performed in a volume of 25μl containing 10XPCR buffer, 500nM of each primer, 2mmol/L MgCl2; 200μM each deoxy nucleotide triphosphate (dNTPs), 1.5U Taq DNA polymerase and 200ng DNA sample. PCR was performed in a thermal cycler (Applied Biosystem 2720) under the following conditions: initial denaturation at 94°C for five minutes was followed by 35 cycles of 93°C for 1 minute, 55°C for 30 seconds, 72°C for 1 minute and final extension at 72°C for 10 minutes. The amplified products were analyzed in 2% agarose gel by electrophoresis. Following electro-phoresis, amplified product, the band of the sample was visualized under UV transilluminator. The bands were identified according to their molecular size by comparing with the molecular weight marker (100 bp DNA ladder) loaded in a separate lane. DNA sequencing: To confirm the type and number of EPIYA motif, direct sequencing of cagA 3´ variable region primer based amplified PCR product was performed. Sequencing was performed by Sanger dideoxy sequencing method at the Center for Advanced Research and Sciences, University of Dhaka. For automated DNA sequencing, PCR products were purified by using Thermo Scientific GeneJet PCR Purification Kit prior to cycle sequencing. The purified DNA was quantified and purities were evaluated by spectrophotometer (Nanodrop, ND 1000, Japan). After checking concentration of DNA, six samples were selected for sequencing. Purified products were sequenced using a BigDye Terminator Cycle Sequencing Kit in an ABI 3130 Genetic Analyzer (Applied Biosystems, USA). The sequences obtained were aligned using the CAP3 Sequence Assembly Program. After alignment, nucleotide sequences were transformed into amino acid sequences using the Blastx program and compared to previously published CagA gene sequence of strains H. pylori 26695 (AE000511) deposited into the GenBank.15 Statistical Analysis: After collection all data were checked, edited and analyzed by using computer based SPSS (Statistical package of social science) software. p value was calculated using Chi-square test to find the significant relationship. p value less than 0.05 was statistically significant.
Result:
Among 78 study population, 56 (71.8%) were male and 22(28.2%) were female with the mean age 39.38±12.82 years (range 18-65 years). Of them majority (51, 65.4%) were diagnosed as gastritis followed by 11(14.1%) duodenal ulcer, 10(12.8%) gastric ulcer and 6(7.7%) gastric carcinoma. Out of 78 gastric biopsy specimens, 31(39.7%) were confirmed to have gastric H. pylori infection by rapid urease test (RUT) and ureC gene. Among 31 H. pylori positive cases, 19(61.3%) were cagA gene positive including 55%(11/20) of gastritis cases, 66.7%(2/3) of gastric ulcer cases, 66.7%(4/6) of duodenal ulcer cases and 100%(2/2) gastric carcinoma cases (table I). cagA gene is significantly associated with duodenal ulcer cases (p=<0.05).
Distribution of cagA EPIYA motif in H. pylori cagA positive cases and their relation to gastroduodenal diseases are shown in table II.
Among 19 cagA positive cases, EPIYA typing PCR showed that majority of gastroduodenal cases (57.9%) had 3 copies of EPIYA (ABC type), 26.3% had 4 copies (ABCC type), while remaining 10.5% had AC and 5.2% had AB type EPIYA motif. Most (75%) of the duodenal ulcer cases composed of ABC type EPIYA motif while 50% of gastric ulcer and gastric carcinoma cases had ABCC type EPIYA motif. There was no East Asian type cagA EPIYA ABD motif.
Nucleotide sequencing of the cagA 3' variable regions was performed for 6 selected cases including 2 from gastritis, 1 from gastric ulcer, 2 from duodenal ulcer and 1 from gastric carcinoma patients. Sequence analyses confirmed that three types of EPIYA motifs were observed: EPIYA-A for EPIYAKVNKKK (A/T/V/S) GQ; EPIYA-B for EPIY (A/T) (Q/K)VAKKVNAKI; and EPIYA-C for EPIYATIDDLGGPFPL (figure 2).
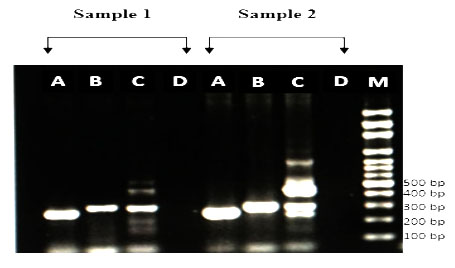
Figure 1: Amplification of cagA EPIYA motif by PCR from H. pylori strains. (1) ABC motifs and (2) ABCC motifs. M: 100-bp DNA marker.
No East Asian type of EPIYA-D (EPIYATIDFDEANQAG) was found. So the present study confirmed that PCR methods correctly classified the EPIYA motif types (i.e. strain-14, 23, 61, 62, 72 possessed ABC type, and strain-77 possessed ABCC type by PCR). The data are identical to data by sequencing.
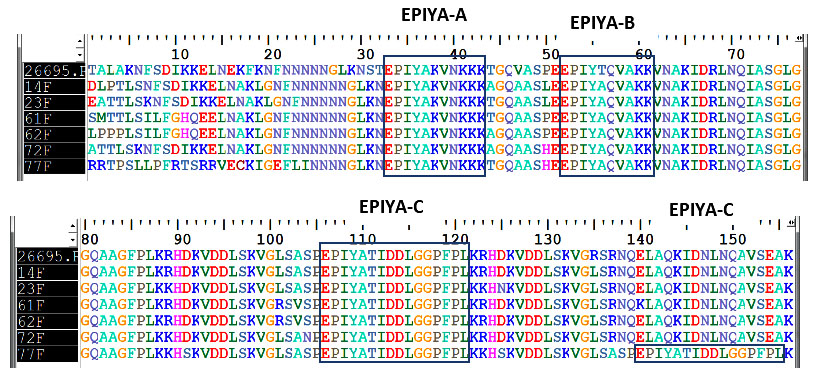
Figure 2: Alignment of amino acid sequences among cagA strain (showing the EPIYA motifs) from 6 H. pylori strains including the H. pylori reference strain 26695.
In this study, out of 31 H. pylori positive cases, 19(61.3%) were cagA gene positive. This finding is consistent with the findings of previous studies showing cagA positivity rate 61% in china, 65.9% in Brazil and 68.4% in Bangladesh.12,15,16
In the present study cagA gene was positive in 66.7% cases in each of duodenal ulcer and gastric ulcer patients compared to 55% cases of gastritis. CagA gene was significantly associated with duodenal ulcer cases (p<0.05). This finding is in agreement with studies done in Japan and Cuba but in contrast to a study done in Singapore reported that there was no association between cagA status and duodenal ulcer.17-19 For this difference in the cagA status, one possibility is the large genomic variations in the H. pylori genomes that amplifies the cagA gene from H. pylori isolated in one country failed to detect cagA in isolates from another country.
Western type cagA EPIYA-C motif was found in all 19 cagA positive H. pylori cases but there was no East Asian cagA EPIYA-D motif. Similar findings were reported from India and Colombia.11,20 In contrast, these results differed from that of Korea where the East Asian type cagA (EPIYA-D) is dominant and some other countries like Malaysia and Thailand where both types of cagA (Western and East Asian) are present.21-23
Among 19 cagA positive cases majority (11,57.9%) were EPIYA ABC motif followed by 5(26.3%) EPIYA ABCC, 2(10.5%) AC and 1(5.2%) AB motif. These results are comparable with the results of previous study done in Colombia.24 The multiple numbers of EPIYA motifs, especially EPIYA-C, is thought to be related to the development of gastro-duodenal diseases. CagA positive H. pylori strains with multiple EPIYA-C motifs are reported to be associated with a higher gastric cancer risk than strains with only one EPIYA-C motif.8,25 In the present study more than one EPIYA-C (ABCC) was found in 26.3% of cagA positive cases which is comparable to the findings reported in South Africa and Colombia.24,26 Geographic variations in the frequency of H. pylori strains with one or two EPIYA-C repeats may be explained by different acidic conditions in the gastric mucosa, which could be related to differences among populations in the frequencies of cytokine gene polymorphisms that attenuate gastric acid secretions.27
Present study showed that the variation in the EPIYA motifs in CagA protein was not directly associated with the outcome of the disease caused by H. pylori and that there is no relation between the number of EPIYA-C motifs and the gastroduodenal diseases (p>0.05). Higher number of EPIYA C segments was associated with gastric carcinoma in Italy and Brazil, otherwise no association was observed in Colombia.24-25 These differences might be due to different study designs, sample size, populations and geographical diversity of H. pylori markers of pathogenicity.
Conclusion
This study shows the pattern of cagA EPIYA motif in gastric biopsy specimen from patients with gastroduodenal diseases. Possibly this is the first study in Bangladesh that shows Western type cagA EPIYA motif was found in all 19 cagA positive H. pylori cases and there is no East Asian EPIYA motif. This finding demand further study to be conducted with more clinical specimens and phylogenetic analysis of the H. pylori genome to determine the ancestry relationship among the overall international isolates. The multiple numbers of EPIYA motifs, especially EPIYA-C, is thought to be related to the development of severe gastro-duodenal diseases and associated with higher gastric cancer risk and was found in 26.3% of cagA positive cases in the present study which also demand more studies to be conducted further to estimate the actual risk of severe gastroduodenal diseases.
Among 78 study population, 56 (71.8%) were male and 22(28.2%) were female with the mean age 39.38±12.82 years (range 18-65 years). Of them majority (51, 65.4%) were diagnosed as gastritis followed by 11(14.1%) duodenal ulcer, 10(12.8%) gastric ulcer and 6(7.7%) gastric carcinoma. Out of 78 gastric biopsy specimens, 31(39.7%) were confirmed to have gastric H. pylori infection by rapid urease test (RUT) and ureC gene. Among 31 H. pylori positive cases, 19(61.3%) were cagA gene positive including 55%(11/20) of gastritis cases, 66.7%(2/3) of gastric ulcer cases, 66.7%(4/6) of duodenal ulcer cases and 100%(2/2) gastric carcinoma cases (table I). cagA gene is significantly associated with duodenal ulcer cases (p=<0.05).
Table I. H. pylori cagA gene among H. pylori positive cases with different gastroduodenal diseases (n=31)
Diseases |
H. pylori positive case |
H. pylori cagA gene positive (%) |
Gastritis |
20 |
11(55) |
Gastric ulcer |
03 |
02(66.7) |
Duodenal ulcer |
06 |
04(66.7) |
Gastric carcinoma |
02 |
02(100) |
Total |
31 |
19(61.3) |
Distribution of cagA EPIYA motif in H. pylori cagA positive cases and their relation to gastroduodenal diseases are shown in table II.
Table II. Distribution of cagA EPIYA motif in H. pylori cagA positive cases in relation to gastroduodenal diseases (n=19)
Diseases |
CagA EPIYA motif (%) |
||||
AB |
AC |
ABC |
ABCC |
ABD |
|
Gastritis(n=11) |
01(9.1) |
02(18.1) |
06(54.5) |
02(18.1) |
00(0) |
Gastric ulcer |
00(0) |
00(0) |
01(50) |
01(50) |
00(0) |
Duodenal ulcer |
00(0) |
00(0) |
03(75) |
01(25) |
00(0) |
Gastric |
00(0) |
00(0) |
01(50) |
01(50) |
00(0) |
Total(n=19) |
01(5.2) |
02(10.5) |
11(57.9) |
05(26.3) |
00(0) |
Among 19 cagA positive cases, EPIYA typing PCR showed that majority of gastroduodenal cases (57.9%) had 3 copies of EPIYA (ABC type), 26.3% had 4 copies (ABCC type), while remaining 10.5% had AC and 5.2% had AB type EPIYA motif. Most (75%) of the duodenal ulcer cases composed of ABC type EPIYA motif while 50% of gastric ulcer and gastric carcinoma cases had ABCC type EPIYA motif. There was no East Asian type cagA EPIYA ABD motif.
Nucleotide sequencing of the cagA 3' variable regions was performed for 6 selected cases including 2 from gastritis, 1 from gastric ulcer, 2 from duodenal ulcer and 1 from gastric carcinoma patients. Sequence analyses confirmed that three types of EPIYA motifs were observed: EPIYA-A for EPIYAKVNKKK (A/T/V/S) GQ; EPIYA-B for EPIY (A/T) (Q/K)VAKKVNAKI; and EPIYA-C for EPIYATIDDLGGPFPL (figure 2).

No East Asian type of EPIYA-D (EPIYATIDFDEANQAG) was found. So the present study confirmed that PCR methods correctly classified the EPIYA motif types (i.e. strain-14, 23, 61, 62, 72 possessed ABC type, and strain-77 possessed ABCC type by PCR). The data are identical to data by sequencing.

Discussion
CagA gene is considered to be a major virulence factor associated with gastric carcinoma. As large numbers of populations in Bangladesh are seropositive for H. pylori so, the present study was done to detect the number and pattern of H. pylori cagA EPIYA motifs using PCR based typing and sequencing analysis to identify the H. pylori infected patients who are at risk to develop gastric carcinoma.In this study, out of 31 H. pylori positive cases, 19(61.3%) were cagA gene positive. This finding is consistent with the findings of previous studies showing cagA positivity rate 61% in china, 65.9% in Brazil and 68.4% in Bangladesh.12,15,16
In the present study cagA gene was positive in 66.7% cases in each of duodenal ulcer and gastric ulcer patients compared to 55% cases of gastritis. CagA gene was significantly associated with duodenal ulcer cases (p<0.05). This finding is in agreement with studies done in Japan and Cuba but in contrast to a study done in Singapore reported that there was no association between cagA status and duodenal ulcer.17-19 For this difference in the cagA status, one possibility is the large genomic variations in the H. pylori genomes that amplifies the cagA gene from H. pylori isolated in one country failed to detect cagA in isolates from another country.
Western type cagA EPIYA-C motif was found in all 19 cagA positive H. pylori cases but there was no East Asian cagA EPIYA-D motif. Similar findings were reported from India and Colombia.11,20 In contrast, these results differed from that of Korea where the East Asian type cagA (EPIYA-D) is dominant and some other countries like Malaysia and Thailand where both types of cagA (Western and East Asian) are present.21-23
Among 19 cagA positive cases majority (11,57.9%) were EPIYA ABC motif followed by 5(26.3%) EPIYA ABCC, 2(10.5%) AC and 1(5.2%) AB motif. These results are comparable with the results of previous study done in Colombia.24 The multiple numbers of EPIYA motifs, especially EPIYA-C, is thought to be related to the development of gastro-duodenal diseases. CagA positive H. pylori strains with multiple EPIYA-C motifs are reported to be associated with a higher gastric cancer risk than strains with only one EPIYA-C motif.8,25 In the present study more than one EPIYA-C (ABCC) was found in 26.3% of cagA positive cases which is comparable to the findings reported in South Africa and Colombia.24,26 Geographic variations in the frequency of H. pylori strains with one or two EPIYA-C repeats may be explained by different acidic conditions in the gastric mucosa, which could be related to differences among populations in the frequencies of cytokine gene polymorphisms that attenuate gastric acid secretions.27
Present study showed that the variation in the EPIYA motifs in CagA protein was not directly associated with the outcome of the disease caused by H. pylori and that there is no relation between the number of EPIYA-C motifs and the gastroduodenal diseases (p>0.05). Higher number of EPIYA C segments was associated with gastric carcinoma in Italy and Brazil, otherwise no association was observed in Colombia.24-25 These differences might be due to different study designs, sample size, populations and geographical diversity of H. pylori markers of pathogenicity.
Conclusion
This study shows the pattern of cagA EPIYA motif in gastric biopsy specimen from patients with gastroduodenal diseases. Possibly this is the first study in Bangladesh that shows Western type cagA EPIYA motif was found in all 19 cagA positive H. pylori cases and there is no East Asian EPIYA motif. This finding demand further study to be conducted with more clinical specimens and phylogenetic analysis of the H. pylori genome to determine the ancestry relationship among the overall international isolates. The multiple numbers of EPIYA motifs, especially EPIYA-C, is thought to be related to the development of severe gastro-duodenal diseases and associated with higher gastric cancer risk and was found in 26.3% of cagA positive cases in the present study which also demand more studies to be conducted further to estimate the actual risk of severe gastroduodenal diseases.
References
- Peek RM and Crabtree JE. Helicobacter infection & gastric neoplasia. J. Pathology. 2006; 208:233-48.
- Ahmad MM, Rahman M, Rumi AK, Islam S, Hug F, Chowdhury MF et al. Prevalence of Helicobacter pylori in asymptomatic population a pilot serological study in Bangladesh. J of Epidemiology. 1997;7: 251-54.
- Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G et al. Molecular characterization of the 128-kDa immune dominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA. 1993; 90: 5791-95.
- Yamaoka Y, Orito E, Mizokami M, Gutierrez O, Saitou N, Kodama T. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 2002; 517:180–184.
- Kim SY, Woo CW, Lee YM, Son BR, Kim JW, Chae HB et al. Genotyping CagA, VacA subtype, IceA1, and BabA of Helicobacter pylori isolates from Korean patients, and their association with gastro-duodenal diseases. J Korean Med Sci. 2001;16:579-84.
- Yamazaki S, Yamakawa A, Okuda T, Ohtani M, Suto H, Ito Y et al. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J. Clin. Microbiol. 2005; 43:3906–16.
- Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002; 295: 683-86.
- Jones K, Joo Y, Jang H, Yoo Y, Lee H, Chung I et al. Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. J. Clin. Microbiol. 2009; 47:959-68.
- Sgouras DN, Panayotopoulou EG, Papadakos K, Martinez-Gonzalez B, Roumbani A, Panayiotou J et al. CagA and VacA polymorphisms do not correlate with severity of histopathological lesions in Helicobacter pylori-infected Greek children. J Clin Microbiol. 2009; 47: 2426-34.
- Naito M, Yamazaki T, Tsutsumi R, Higashi H, Onoe K, Yamazaki S et al. Influence of EPIYA-Repeat Polymorphism on the Phosphorylation-Dependent Biological Activity of Helicobacter pylori CagA. Gastroenterology. 2006; 130: 1181-90.
- Tiwari SK, Sharma V, Sharma VK, Gopi M, Saikant R, Nandan A et al. Phylogenetic analysis, based on EPIYA repeats in the cagA gene of Indian Helicobacter pylori, and the implications of sequence variation in tyrosine phosphorylation motifs on determining the clinical outcome. Genetics and Molecular Biology. 2011 (cited 2014 Dec 21); 34, 2, 280-85. Available from: http://dx.doi.org/10.1 590/S1415-47572011005000003.
- Rahman M, Mukhopadhyay AK, Nahar S, Datta S, Ahmad MM, Sarkar S et al. DNA-level characterization of Helicobacter pylori strains from patients with overt disease and with benign infections in Bangladesh. J Clin Microbiol. 2003; 41:2008-14.
- Rahman SHZ, Rahman MA, Arfin MS, Alam MM, Bhuiyan TM, Ahmed N et al. Helicobacter pylori Infection and Strain Types in Adult Dyspeptic Patients Undergoing Endoscopy in a Specialized Hospital of Dhaka City. Bangladesh J Med Microbiol. 2009; 03: 4-9.
- Lu JJ, Perng CL, Shyu RY, Chen CH, Lou Q, Chong SK, et al. Comparison of Five PCR Methods for Detection of Helicobacter pylori DNA in Gastric Tissues. J Clin Microbiol. 1999; 37: 772- 74.
- Silva AV, Silva MR Junior, Vinagre RMDF, Santos AK, Costa RAA, Fecury AA et al. Evaluation of the Pattern of EPIYA Motifs in the Helicobacter pylori cagA Gene of Patients with Gastritis and Gastric Adenocarcinoma from the Brazilian Amazon Region International Journal of Bacteriology.2014 (cited 2014 Dec 25). Available from http://dx.doi.org/10.1155/2014/ 418063.
- Aziz F, Chen X, Yang X and Yan Q. Prevalence and correlation with clinical disease of the Helicobacter pylori cagA and vacA genotype among gastric patients from northeast China. Biomed Research International. 2014 (cited 2014 Dec 15). Available at http://dx.doi.org /10.1155/2014/14298.
- Yamaoka Y, El-Zimaity HM, Gutierrez O, Figura N, Kim JC, Kodama T et al. Relationship between the cagA 3' repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterology. 1999; 117:342-349.
- Torres L, Melián K, Moreno A, Alonso J, Sabatier C, Harnandez M et al. Prevalence of vacA, cagA and babA2 genes in Cuban Helicobacter pylori isolates. World J Gastroenterol. 2009; 15:204-10.
- Zheng PY, Hua J, Yeoh KG, and Ho B. Association of peptic ulcer with increased expression of Lewis antigens but not CagA, iceA, and VacA in Helicobacter pylori isolates in an Asian population. Gut. 2000; 47: 18-22.
- Sicinschi LA, Correa P, Peek RM, Camargo MC, Piazuelo MB, Romero-Gallo J et al. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2010; 16:369-78.
- Choi KD, Kim N, Lee DH, Kim JM, Kim JS, Jung HS, et al. Analysis of the 3'variable region of the cagA gene of Helicobacter pylori isolated in Koreans. Dig.Dis.Sci. 2007; 52:960-66.
- Schmidt H, Andres S, Kaakoush N, Engstrand L, Eriksson L, Goh K, et al. The prevalence of the duodenal ulcer promoting gene (dupA) in Helicobacter pylori isolates varies by ethnic group and is not universally associated with disease development: a case-control study. Gut Pathog. 2009; 1:5.
- Chomvarin C, Phusri K, Sawadpanich K, Mairiang P, Namwat W, Wongkham C et al. Prevalence of cagA EPIYA motifs in Helicobacter pylori among dyspeptic patients in Northeast Thailand. SoutheastAsian J Trop Med Public Health. 2012; 43:446-49.
- Acosta N, Quiroga A, Delgado P, Bravo MM. Helicobacter pylori CagA protein polymorphisms and their lack of association with pathogenesis. World J Gastroenterol.2010; 16:3936-43.
- Yamaoka Y, Osato MS, Sepulveda AR, Gutierrez O, Figura N, Kim JG et al. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol Infect. 2000; 124: 91-96.
- Basso D, Zambon CF, Letley DP, Stranges A, Marchet A, Rhead JL et al. Clinical Relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008; 135:91-99.
- Snaith A and El-Omar EM. Helicobacter pylori Host genetics and disease outcomes. Expert rev Gastroenterology Hepatol. 2008; 2:577-85.